library(igraph) # Networks
library(ADAPTSNA) # Data6 Cleaning Network Data - Trimming and Adding
This script is intended to help you to clean up network data that you have collected or got access to.
| LEARNING ELEMENTS - Data Practices |
|
6.1 Deleting Nodes.
To delete nodes from your network, you use the delete_vertices() function in igraph. There are several reasons you may wish to remove nodes from the network. One very common issue with cleaning network data is knowing what to do with nodes isolates. Isolates are those who are a part of your network, but who have no connections to others in the group. Isolates are stored in network data differently depending on how your data are stored.
If your data are stored in an adjacency matrix, then isolates are those with no 1s in the matrix. Ensuring that R recognises them as isolated is very simple. Bring in the data, and then convert it into a matrix. Any that are isolated will show as isolates. The process of deleting or adding nodes and edges apply to any network object either made from an edgelist or adjacency matrix.
However, dealing with isolates is not as straightforward when you are working with edgelists. With this structure, you have only two columns, one for senders and the other for receivers. If there is an individual in the group who neither sends nor receives, but is a legitimate participant of the group, what do you do with them? One way of recording such isolates in an edgelist is list them as connected to themselves (known as a self loop). Take a look at this edgelist and you will see that these individuals are connected to themselves.
hog_crush_loops <- load_data("Hogwarts Crushes Edgelist_SELFLOOPS.csv", header=TRUE)
#Take a look at the data
hog_crush_loops Crusher Crush
1 Harry Potter Ginny Weasley
2 Harry Potter Cho Chang
3 Ron Weasley Hermione Granger
4 Hermione Granger Ron Weasley
5 Ron Weasley Lavender Brown
6 Ginny Weasley Harry Potter
7 Lily Potter James Potter
8 James Potter Lily Potter
9 Severus Snape Lily Potter
10 Nymphadora Tonks Remus Lupin
11 Remus Lupin Nymphadora Tonks
12 Lavender Brown Ron Weasley
13 Cho Chang Cedric Diggory
14 Cho Chang Harry Potter
15 Cedric Diggory Cho Chang
16 McGonagal McGonagal
17 Madeye Madeye
18 Voldemort Voldemort
19 Flitwick FlitwickNow when you make this a graph object R does something different.
Crush_loops <- graph_from_data_frame(hog_crush_loops, directed = TRUE)
plot(Crush_loops)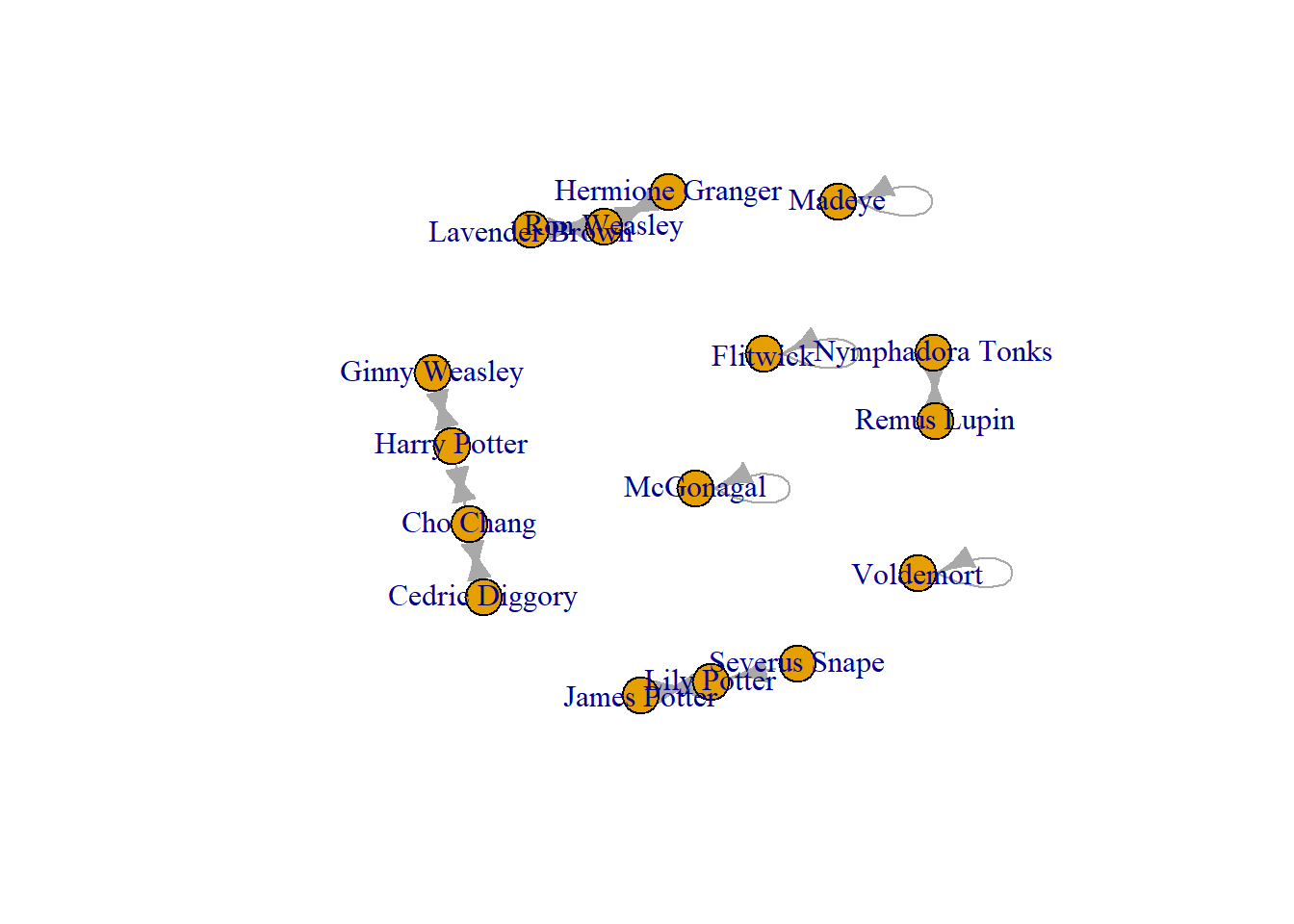
These do not look great, and can cause confusion to viewers of the network visual and even influence some of the mathematics of your analysis. We will deal with those in a moment (next step deleting edges).
Another way to record isolates from an edgelist is to list no-one in the “to” column. In other words, you list the name of the person in your network but leave the cell next to them blank. However, this approach also has additional steps to take before it is clean and ready to go.
hog_crush_empty <- load_data("Hogwarts Crushes Edgelist_EMPTY.csv", header=TRUE)Take a look at the edgeist now it is in and you will see I added a few more characters to this group: Madeye, Flitwick, McGonagal, and Voldemort. They are all listed in the “Crusher” (from) column but have no connection to anyone in the “crush” column. This makes sense, since we know little about their romances from the Harry Potter Saga.
hog_crush_empty Crusher Crush
1 Harry Potter Ginny Weasley
2 Harry Potter Cho Chang
3 Ron Weasley Hermione Granger
4 Hermione Granger Ron Weasley
5 Ron Weasley Lavender Brown
6 Ginny Weasley Harry Potter
7 Lily Potter James Potter
8 James Potter Lily Potter
9 Severus Snape Lily Potter
10 Nymphadora Tonks Remus Lupin
11 Remus Lupin Nymphadora Tonks
12 Lavender Brown Ron Weasley
13 Cho Chang Cedric Diggory
14 Cho Chang Harry Potter
15 Cedric Diggory Cho Chang
16 McGonagal
17 Madeye
18 Voldemort
19 Flitwick When we make this a graph object, R does something funky.
The new characters are all connected to a nameless node and it looks, on visual inspection, that they all have a crush on the same person.
I have highlighted that node in the visualization below. The red node is nameless because the edgelist has empty (nameless) cells.
crush_empty <- graph_from_data_frame(hog_crush_empty, directed = TRUE)
plot(crush_empty)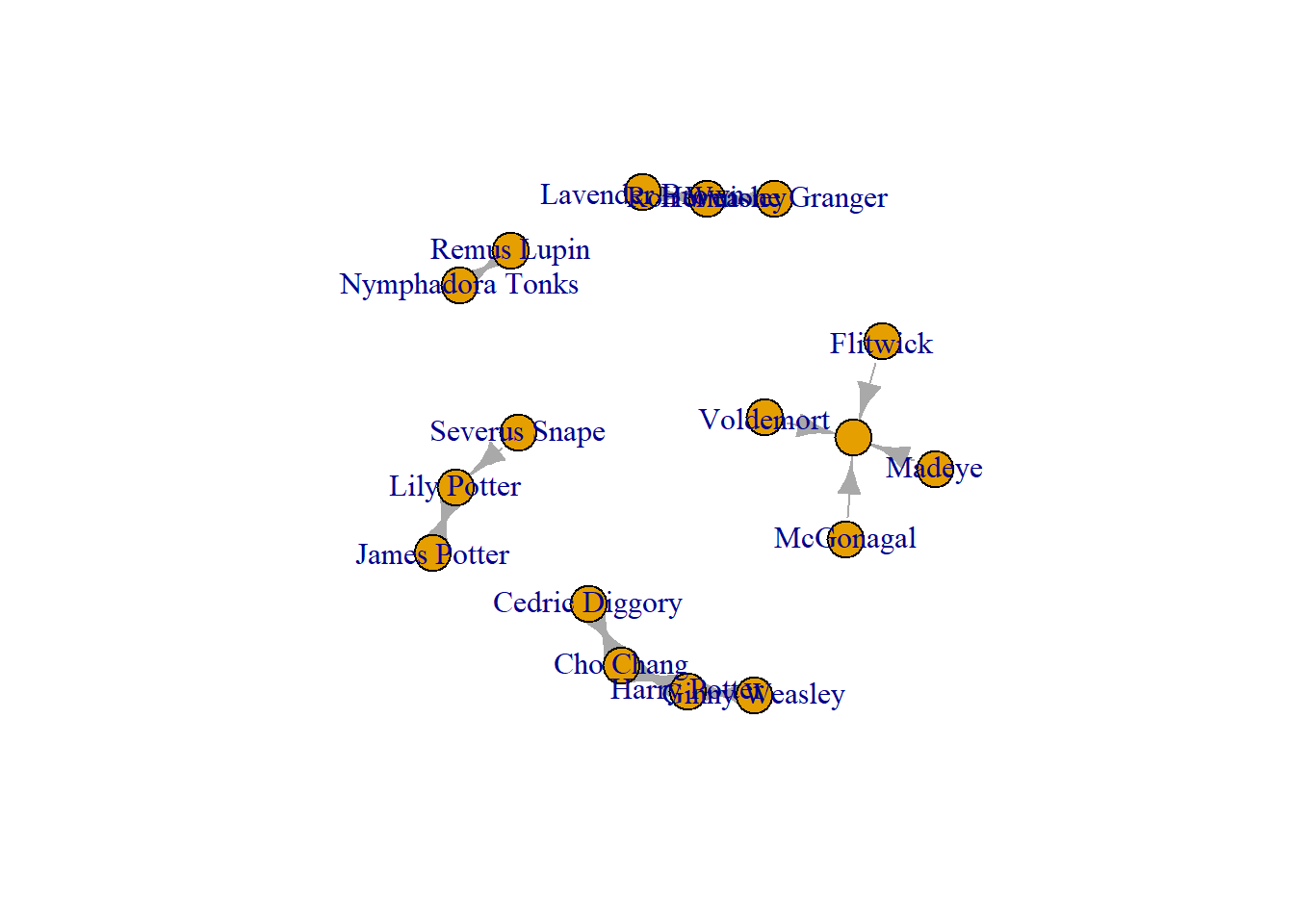
V(crush_empty)$wrong <- ifelse(V(crush_empty)$name %in% c(""), "red", "white")
plot(crush_empty, vertex.color = V(crush_empty)$wrong)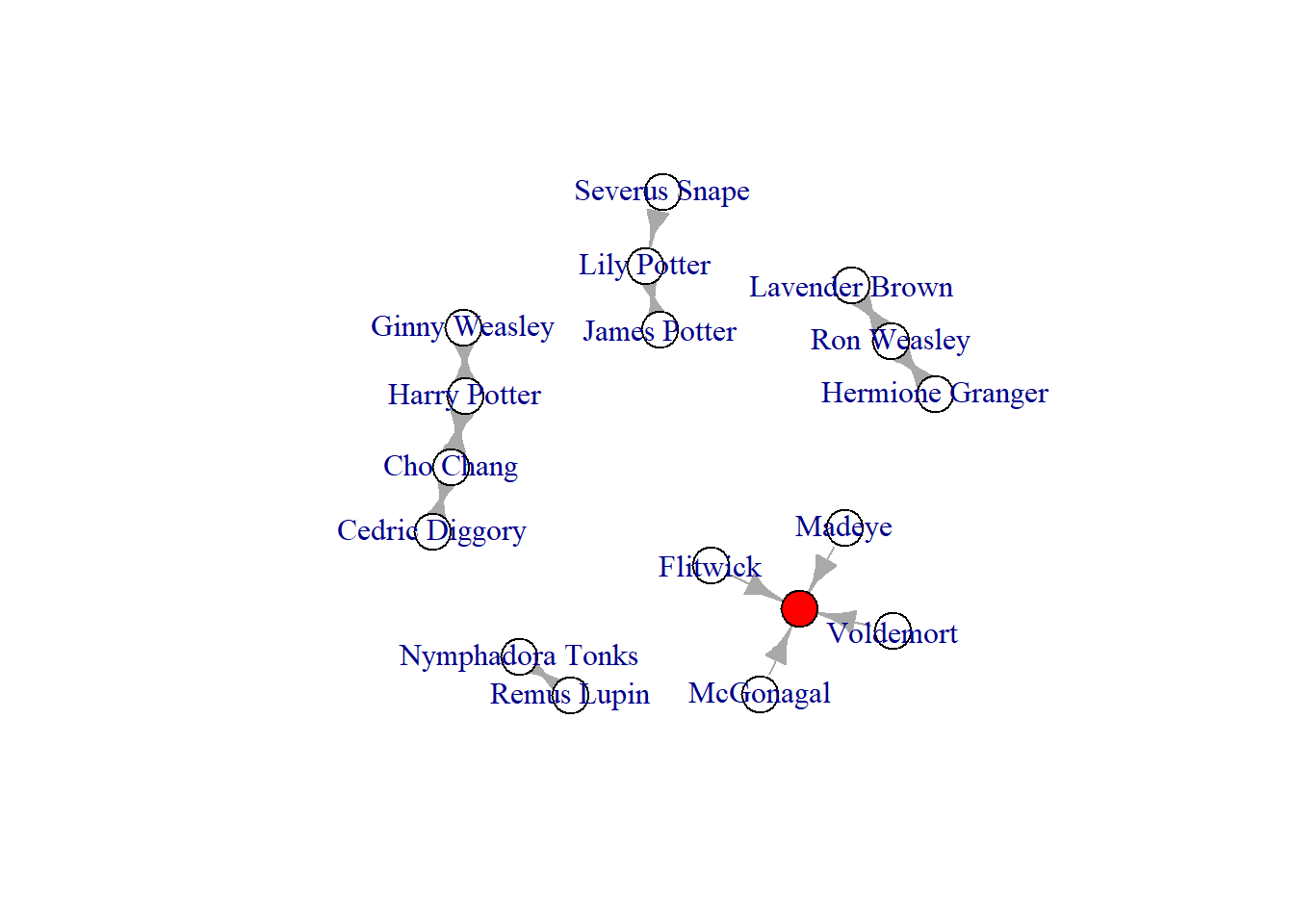
One way to deal with this is to delete the superfluous node. You do this using the delete_vertex() function. ##This fixes the issue once you have the data in Rstudio, but the issue still exists in your dataset. If you choose to structure your network data this way, you will have to remember to remove this node every time. This may be harder to do/realise when dealing with large dense networks.
crush_empty <- delete_vertices(crush_empty, "")
plot(crush_empty)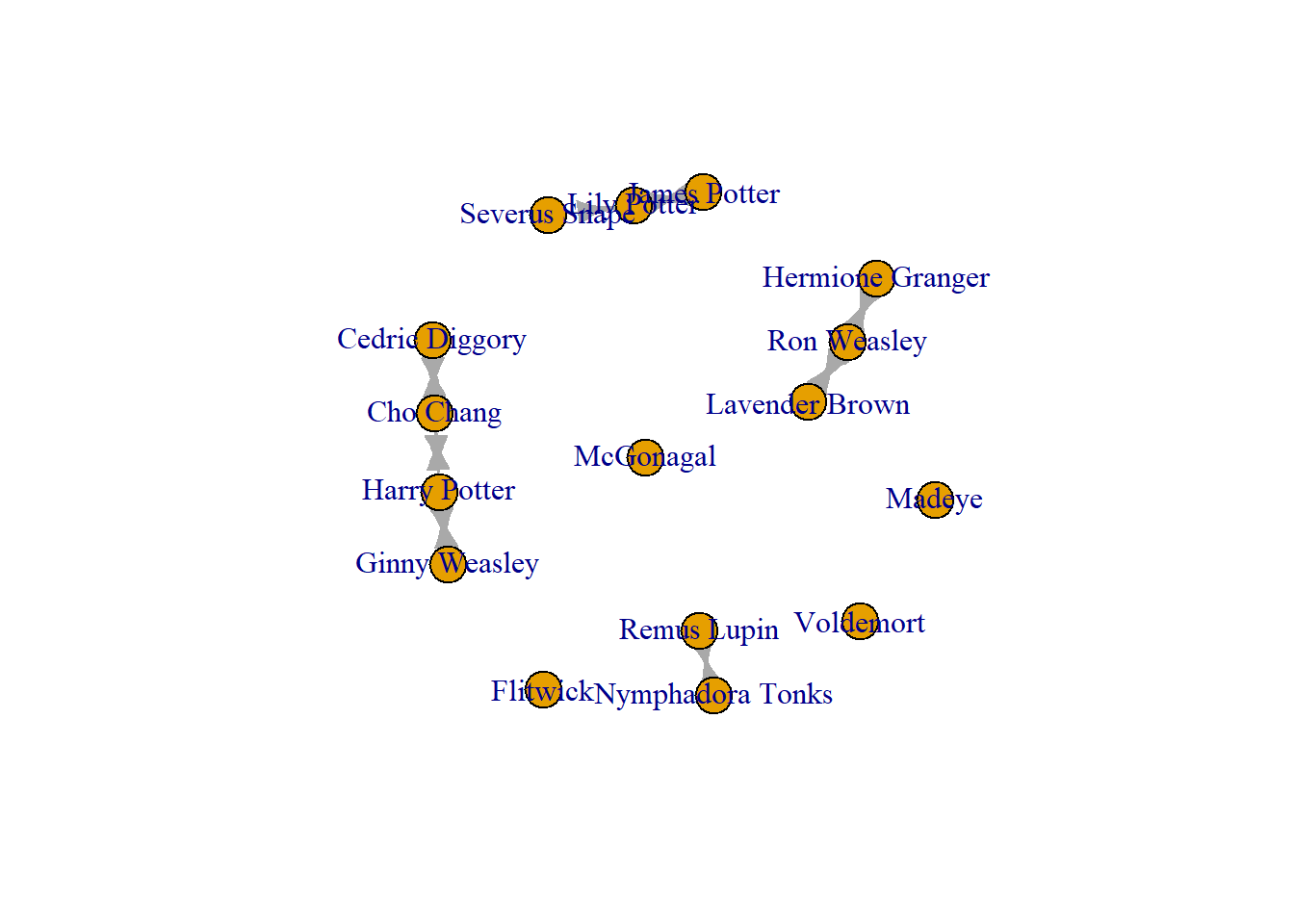
Sometimes, you want to remove all of the isolated nodes from your network because you only care about those who have connections to others. To do this, you identify those with no connections (degree = 0) and them remove them from your network. I suggest making a new object with this sub network.
hog_crush_isol <- which(degree(crush_empty)==0)Now you use the delete_vertices() command and remove those in the vector you just created (those with degree = 0)
Crush_no_isol <-delete_vertices(crush_empty, hog_crush_isol)
plot(Crush_no_isol)
Now this new object has only those nodes with ties to others in the network.
Other than isolates, you you might decide to remove one or more specific nodes from your network. For example, in this hogwarts dataset, we may want to remove those who are not students at Hogwarts (i.e. remove teachers or adults). To do this, you would use the delete_vertices() option.
Another approach is to delete them one-by-one and identify them by their name. Let’s say you have a network and only want to remove one node. You can do so based on their name. To do this, let’s now switch to the other data we brought in, the one with the self loops (we will deal with those next!). We can use the same function, delete_vertices(), but this time, state the name of the node we want to delete.
hog_crush_students <- delete_vertices(Crush_loops, "Voldemort")
plot(hog_crush_students)
A quicker way, if you are deleting multiple, is to make a vector with all the names of those you want to remove, then use the delete_vertices() command.
hog_adults <- c("Severus Snape", "Lily Potter", "James Potter", "Nymphadora Tonks", "Remus Lupin", "Voldemort", "Flitwick", "McGonagal", "Madeye")
hog_crush_students <- delete_vertices(Crush_loops, hog_adults)
plot(hog_crush_students)
This new version removed all unwanted nodes at once.
6.2 Deleting edges
To remove unwanted edges you can use the delete_edges() command. Let’s begin with our edgelist from above and select the edges that are looped by using the E() command coupled with the is.loop() options.
Crush_loops <- delete_edges(Crush_loops, E(Crush_loops )[which_loop(Crush_loops)])
plot(Crush_loops)
In addition to the selfloops, you may want to delete edges between two specific nodes. You can do so by selecting
edges_to_delete <- E(Crush_loops)[(.from("Remus Lupin") & .to("Nymphadora Tonks"))]
Crush_edge_delete <- delete_edges(Crush_loops, edges_to_delete)
plot(Crush_edge_delete)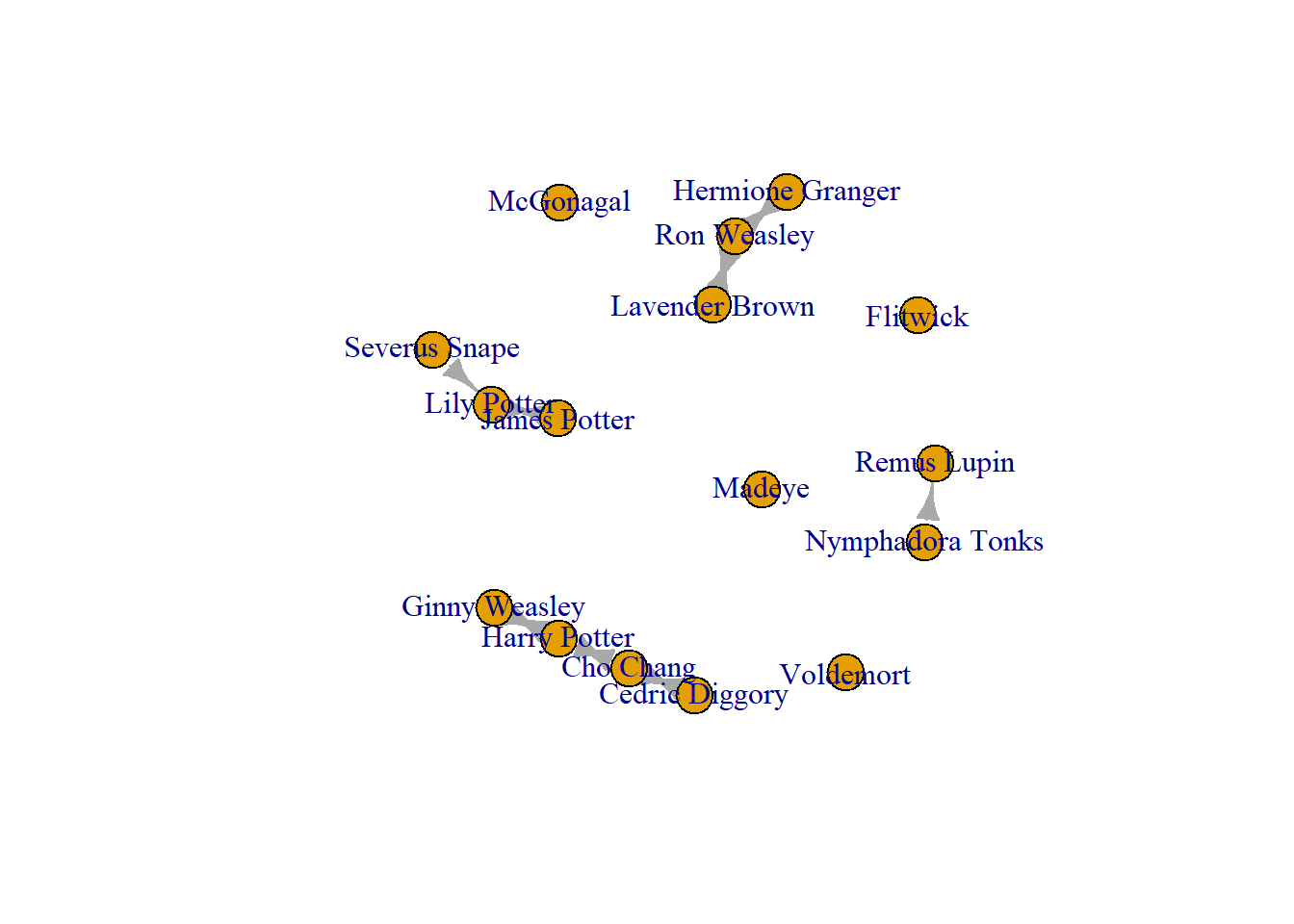
To delete all edges between two nodes.
edges_to_delete2 <- E(Crush_loops)[(.from("Remus Lupin") & .to("Nymphadora Tonks")) | .from("Nymphadora Tonks") & .to("Remus Lupin")]
Crush_edge_delete <- delete_edges(Crush_loops, edges_to_delete2)
plot(Crush_edge_delete)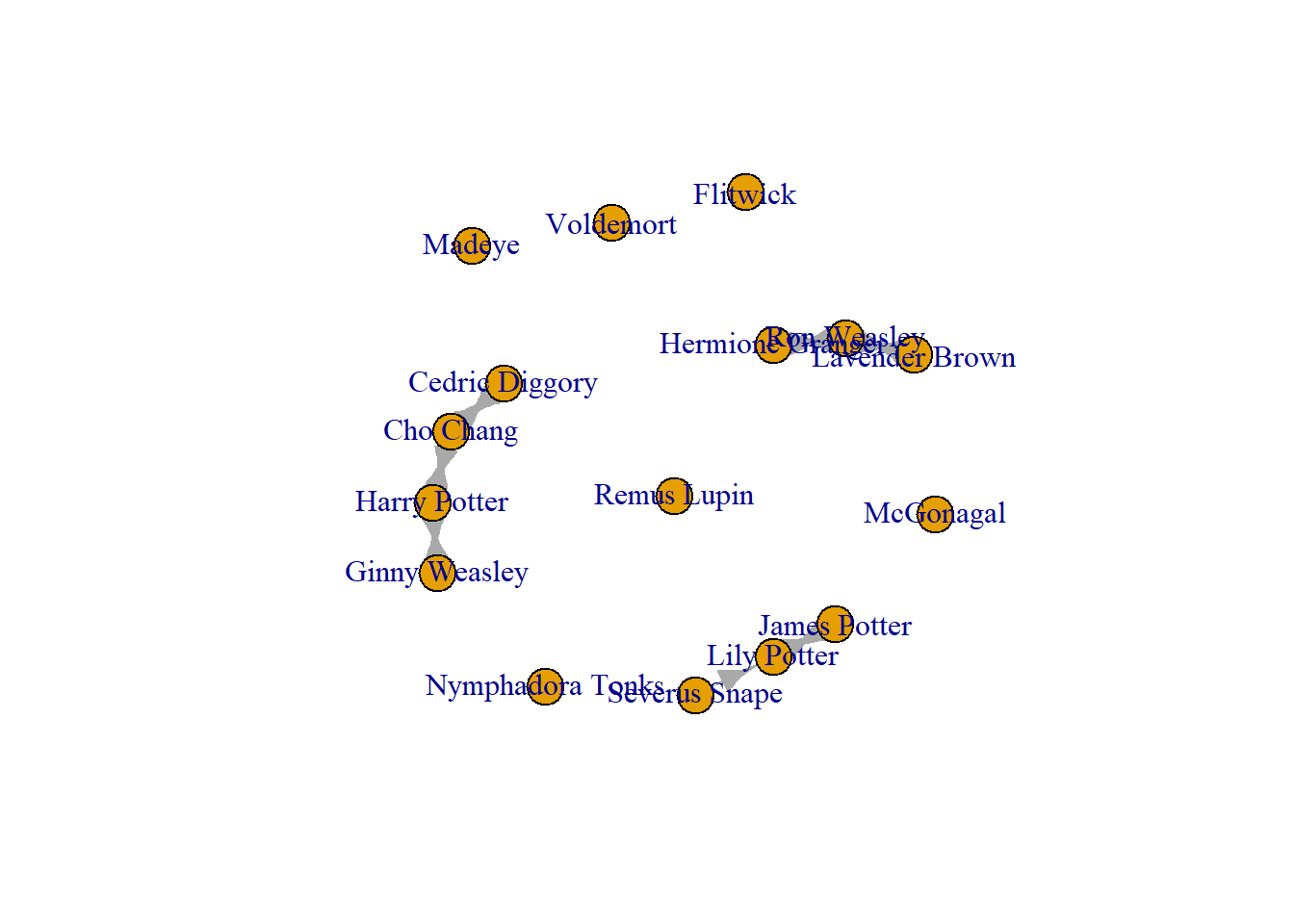
6.3 Adding Nodes
Next, you might need to add data to the network object. Use caution when doing this. Consider the ethics of your research. Has the person you are adding consented to being studied? Adding data, any data, requires consideration.
Once you have considered the above and are sure that it is ethical to add data to your network, you may wish to add nodes to your network. To do so, use add.vertices() function.
crush_added <- add.vertices(Crush_loops, 1, name = "Michael Corner")
plot(crush_added)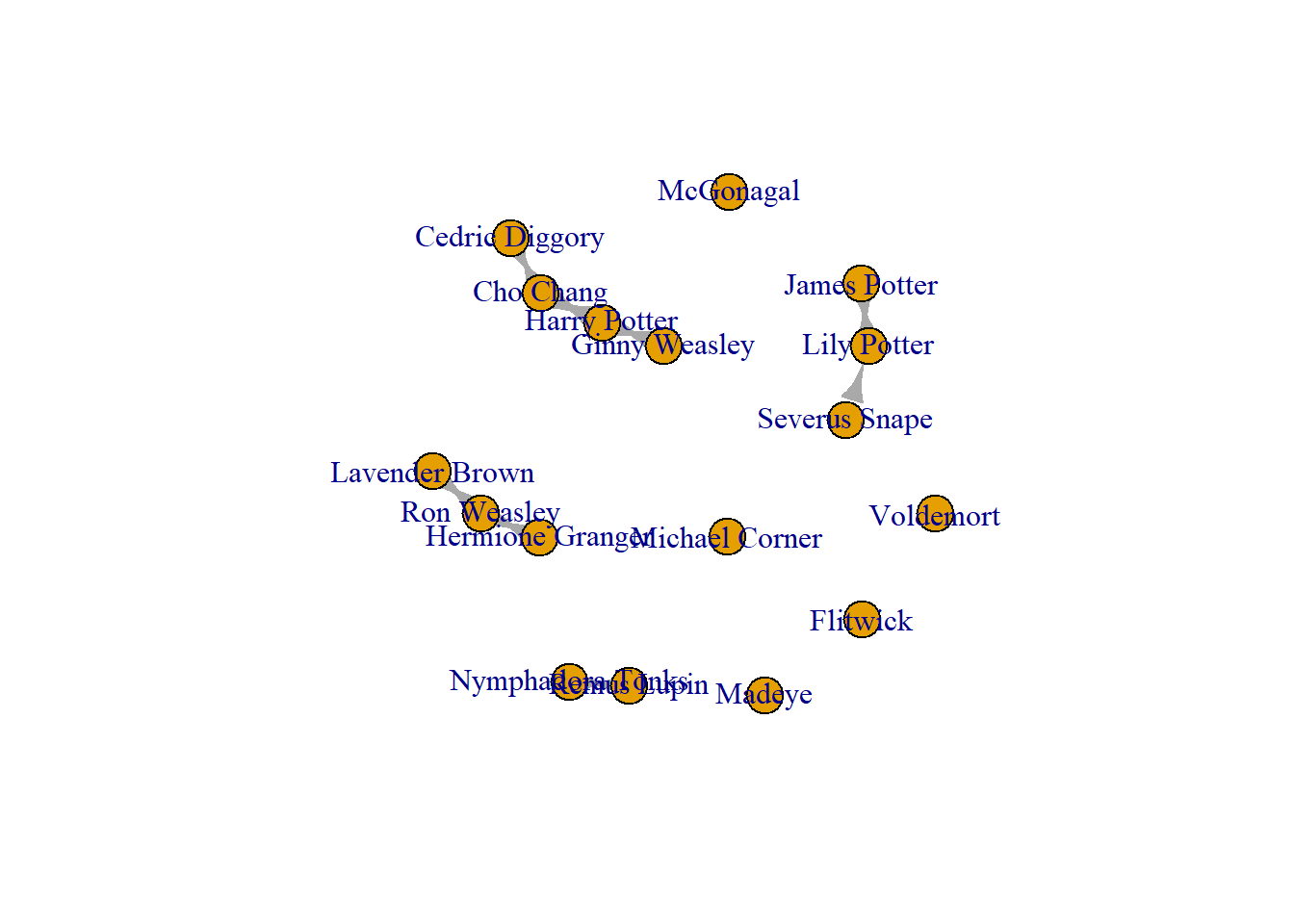
This function follows the following logic: you state the network that you want to add to, state how many nodes you are adding (in this case 1), then state the attribute of the node you are adding (in this case, the name is “Michael Corner.”
6.4 Add Edges
You may need to add edges that you know exist. For example, in this network, the node that we added, “Michael Corner” has connections to another in this network! So, when I originally collected these network data, I forgot this guy! Note, these are fictional people, so I do not need to check with “Michael Corner” before I add him and his connections to our network.
We have added him in, but now we need to add his connections. To do this, you use add.edges().Note, that we start with Michael, then we list to whom he is connected. Think of this as a “from” and “to” formula. So here, this goes from “Michael Corner” to “Ginny Weasley.”
crush_added <- add.edges(crush_added, edges = c("Michael Corner", "Ginny Weasley"))
plot(crush_added)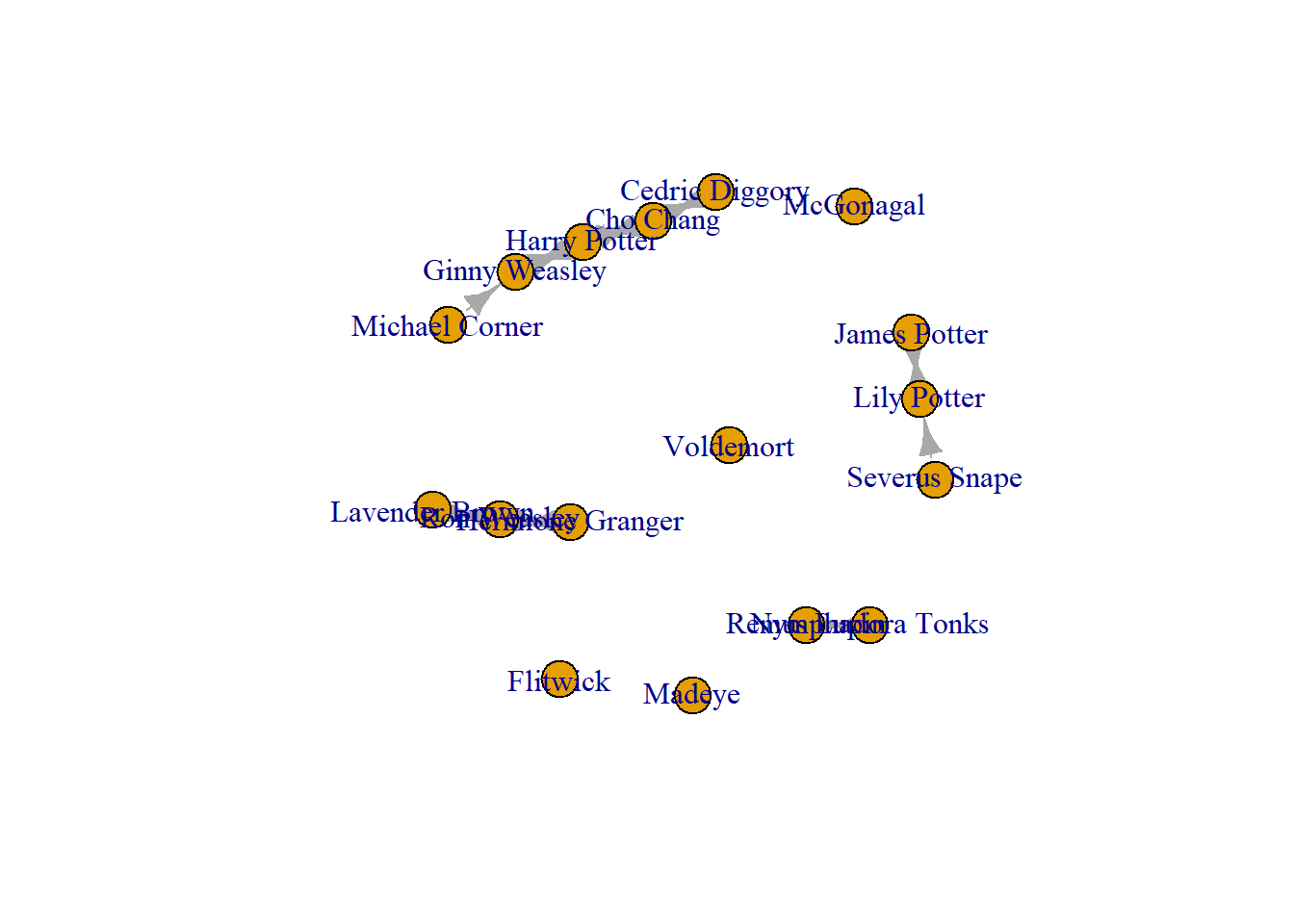
Now to add the reciprocated tie, from “Gunny Wealey” to “Michael Corner.”
crush_added <- add.edges(crush_added, edges = c("Ginny Weasley", "Michael Corner"))
plot(crush_added)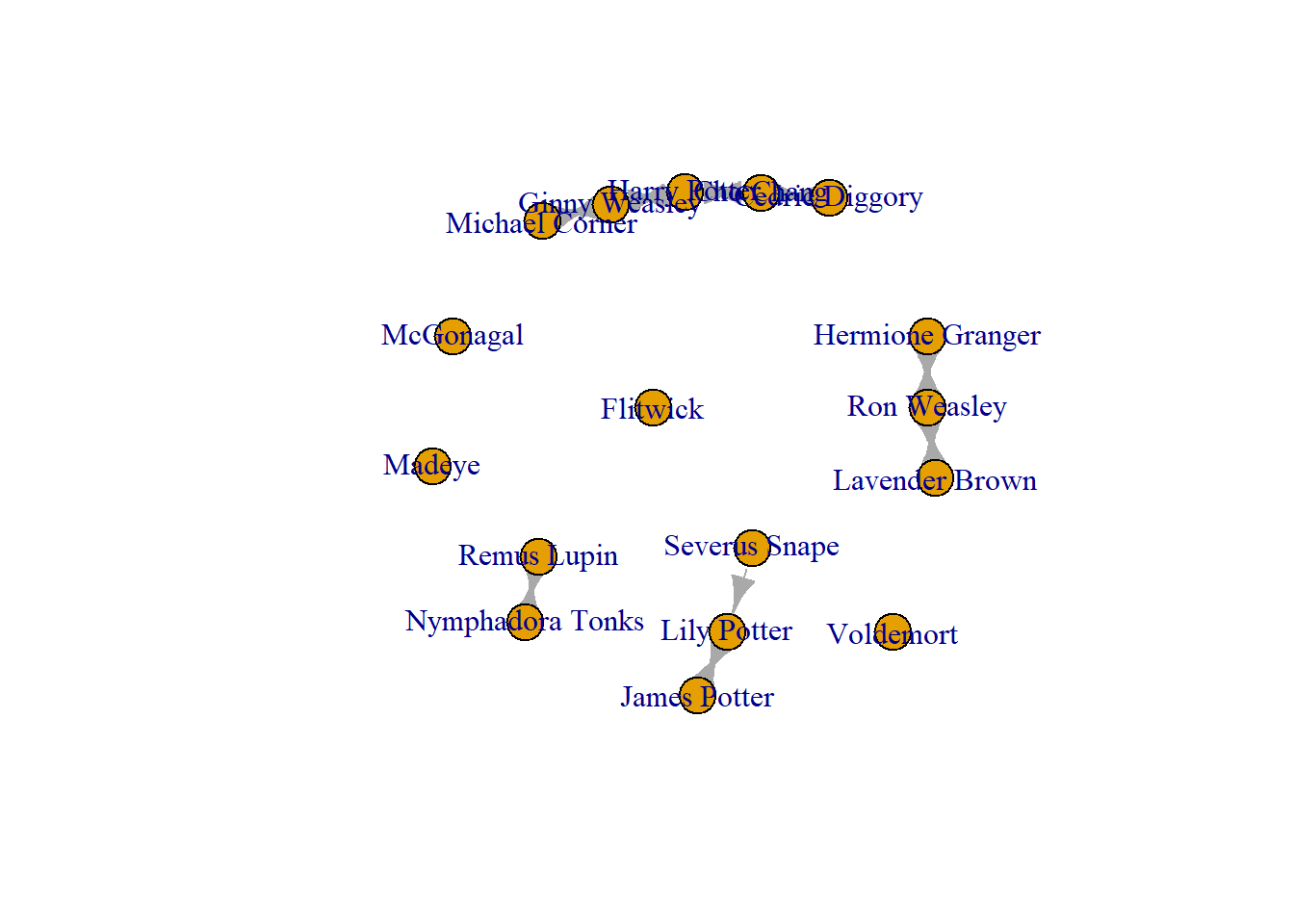
6.5 Summary
Here we have covered a few ways to clean network data. We have covered a few tings.
Network data can be messy
Adding or delete nodes and edges either systematically (i.e. all isolates) or one-by-one.
Well done!
